Research Area
Genetic and infectious diseases remain major threats to human and public health. Yet, current methods and technologies for their treatment and detection are often limited in sensitivity, precision, and accessibility. As a researcher specializing in biological engineering, my academic footprint is firmly rooted in improving human health. With an emphasis on CRISPR system engineering, my research is to develop bioengineering and biosensing approaches that can be applied to treat genetic diseases and diagnose infectious diseases. The innovative technologies developed in my research lab can solve the critical and emerging health challenges. In addition, these technologies are being applied in the fields of agricultural and food systems to improve crop resilience, enhance food safety, and enable rapid detection of plant pathogens.
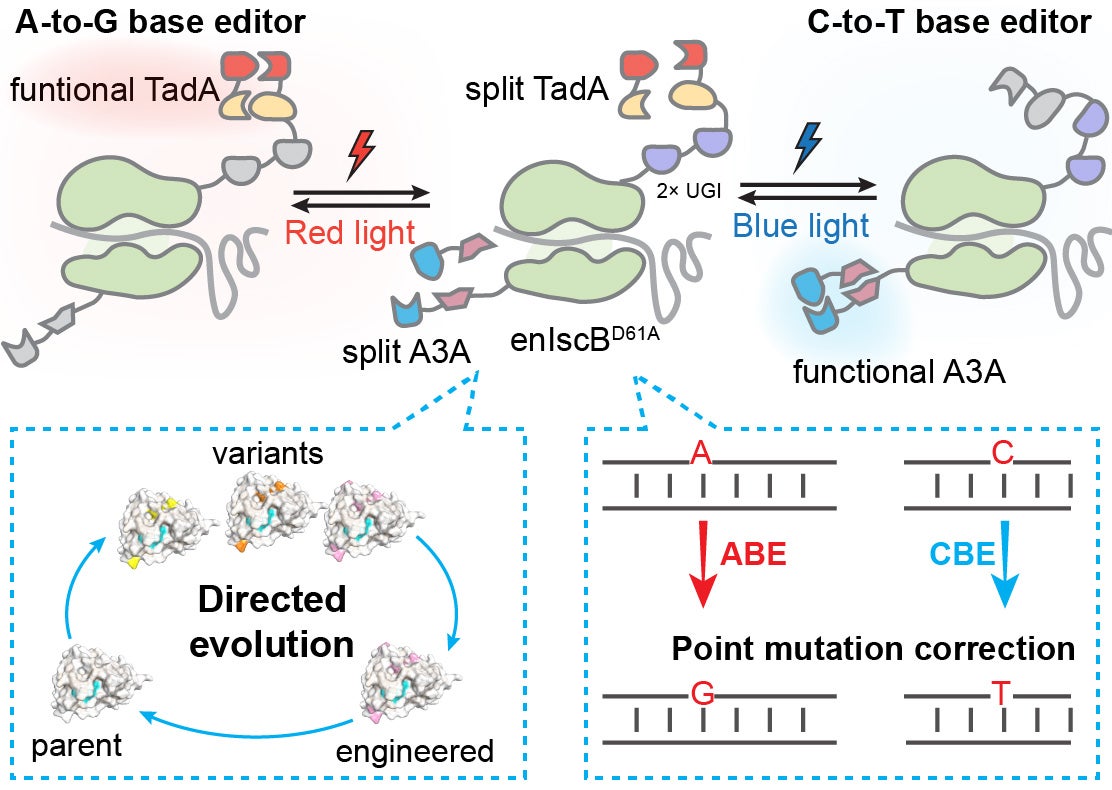
Genome editing
Human diseases, such as metabolic, neurological, and cardiovascular disorders, remain among the leading causes of morbidity and mortality worldwide. They account for millions of deaths each year and place a substantial burden on healthcare systems. The development of many human diseases is highly complex, arising from intricate interactions among genetic, environmental, and lifestyle factors. A notable subset is caused by genetic point mutations that regulate critical biological processes such as metabolism, cellular signaling, or structural protein function. These genetic alterations can disrupt normal physiology, leading to severe dysfunction or death. Traditional treatment strategies, including surgical interventions, pharmacological therapy, or device implantation, mainly alleviate symptoms and slow disease progression, but they cannot address the underlying genetic mutations. This highlights the urgent need for therapeutic approaches that can correct these molecular defects to provide curative solutions.
We are developing base and prime editing approaches using the CRISPR system to precisely and efficiently correct point mutations in genetic diseases, with the goal of restoring normal cellular function at the molecular level.
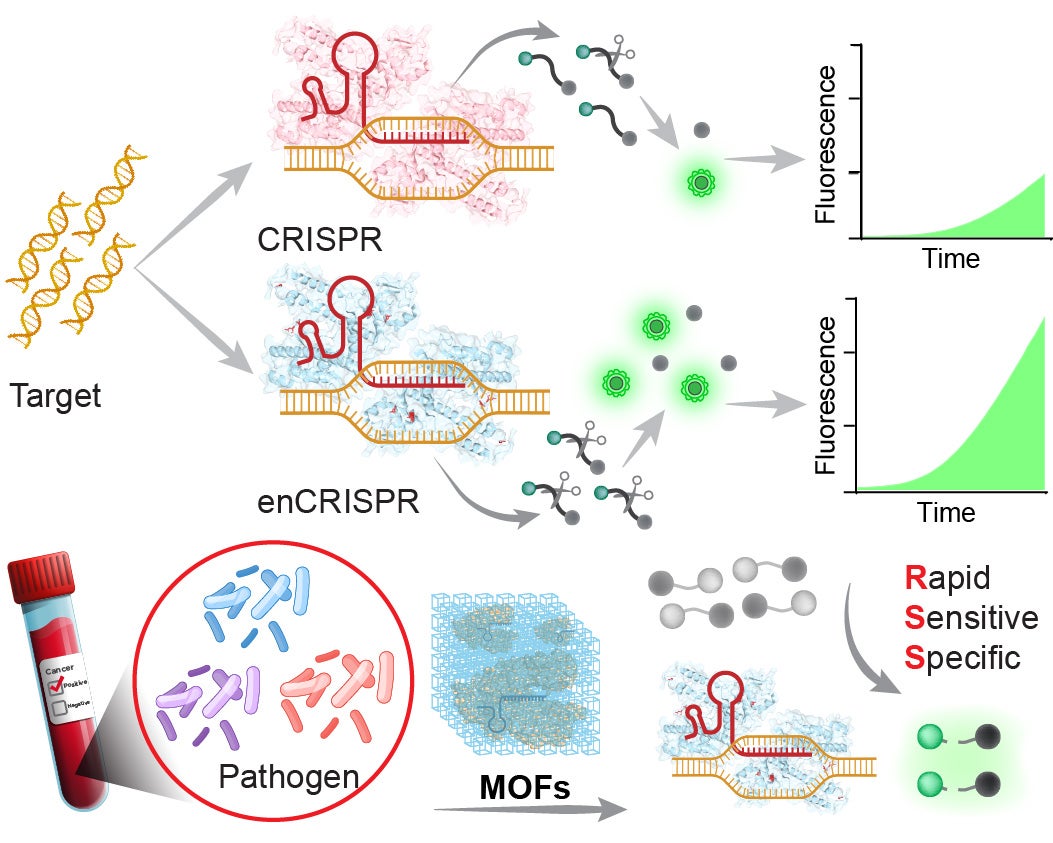
Diagnosis
Infectious diseases remain among the leading causes of morbidity and mortality worldwide and pose a persistent threat to global public health. They account for millions of deaths each year and continue to impose a significant economic and social burden. In the United States alone, infectious diseases contribute substantially to hospitalizations, mortality, and healthcare expenditures. These diseases arise from pathogenic infections that trigger abnormal or uncontrolled immune responses, which can progress to systemic inflammation, multiorgan dysfunction, or even death. The clinical manifestations are highly heterogeneous, reflecting the diversity of causative agents, including bacteria, viruses, fungi, and parasites. A variety of diagnostic methods have been developed to detect infectious diseases in clinical samples, such as immunoassays, molecular diagnostics, and sequencing-based approaches. However, current diagnostic practices often fall short of clinical needs for rapid pathogen detection and risk assessment, which are essential for timely and effective treatment. Therefore, there remains an urgent need to establish rapid, sensitive, and reliable diagnostic tools for infectious diseases.
We are developing CRISPR-based diagnostic assays that integrate sequence-specific recognition with signal amplification as a promising approach to conventional methods for sepsis diagnosis.
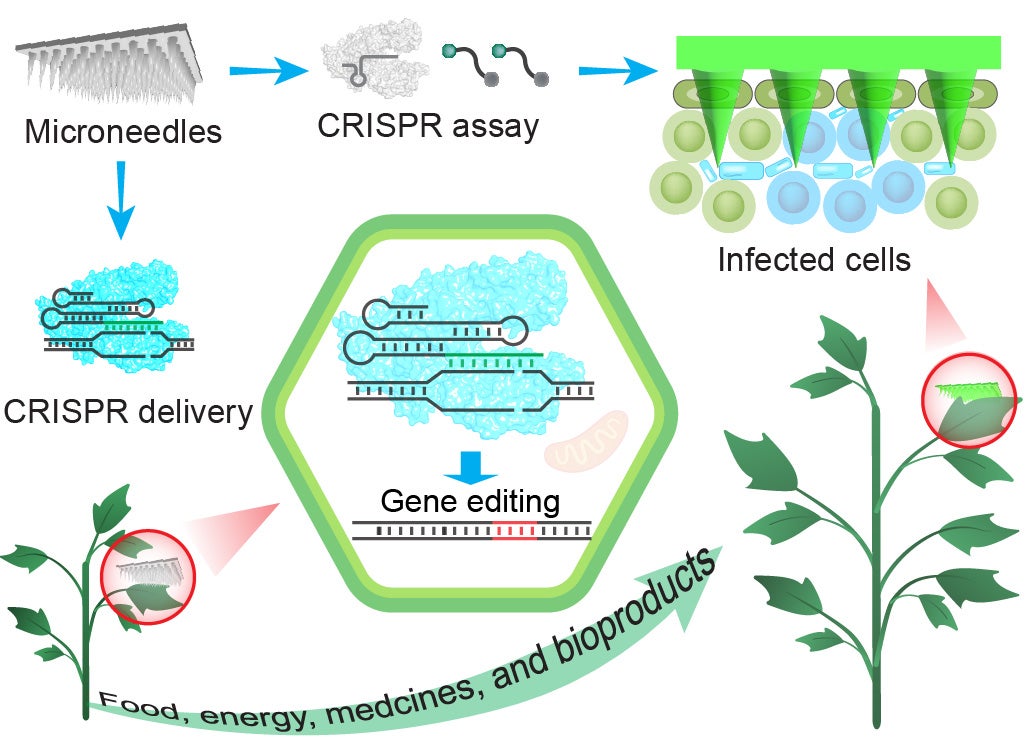
Agriculture
Plants are fundamental to global food security, serving as the primary source of nutrition for humans and livestock. By 2050, global food production must increase by nearly 100% to meet the demands of a rapidly growing population. However, plant diseases pose a major threat to agricultural productivity and food availability. It is estimated that approximately 30% of global crop yields are lost due to plant pathogens. To mitigate these losses, pesticides are extensively applied to control or eliminate pathogens. Yet, the widespread use of pesticides has raised serious environmental and health concerns. On the other hand, early and accurate disease detection is a significant challenge. The existing detection methods rely heavily on laboratory analysis that requires trained personnel, expensive equipment, and complex sample preparation. These limitations hinder rapid and on-site disease detection and make such approaches impractical for field applications. Therefore, there is an urgent need for advanced and innovative technologies that can support sustainable crop management and global food security.
We are applying CRISPR technologies to enhance food production by combining gene editing and molecular detection. Through precise genome editing, we are developing crops with improved yield, nutritional quality, and resistance to diseases, pests, and environmental stresses.